Project organization
Why we need a principled approach to project organization
- helps you structure your work
- helps working faster and safer
- helps reproducibility and collaboration
(My) Best practices
The following is an incomplete list of what I think are good practices for working with R (and other programming projects).
These practices should help making your work more reproducible, intelligible to you and others, and overall easier to work with.
For more, see also the blog series what they forgot to teach you about R
Working with R projects
- Always work within an R project!
- The R.project file should sit in the root folder of your project
Use the same folder structure for all of your projects
Make subfolders for
- data (and also raw and processed data)
- scripts
- … other folders as needed
- If you want to go full on out, you can use project templates when setting up an R project to automatically create folders, files and other structures for you.
Use the same folder structure for all of your projects
This could look something like this:
-- project_directory/
| -- data/
| -- raw/
| -- processed/
| -- scripts/
| -- cleaning/
| -- analysis/
| -- writeup/
-- .gitignore
-- r_project_file.Rproj
For each project, create a readme file, or other kind of documentation
A good readme should include
- description: what is the project about
- contributors: who is working on this project
- requirements: particular dependencies / software that needs to be installed?
- Roadmap: current stage of the project, and planned stuff (e.g. a todo list)
Reproducibility
Always start R with a blank slate
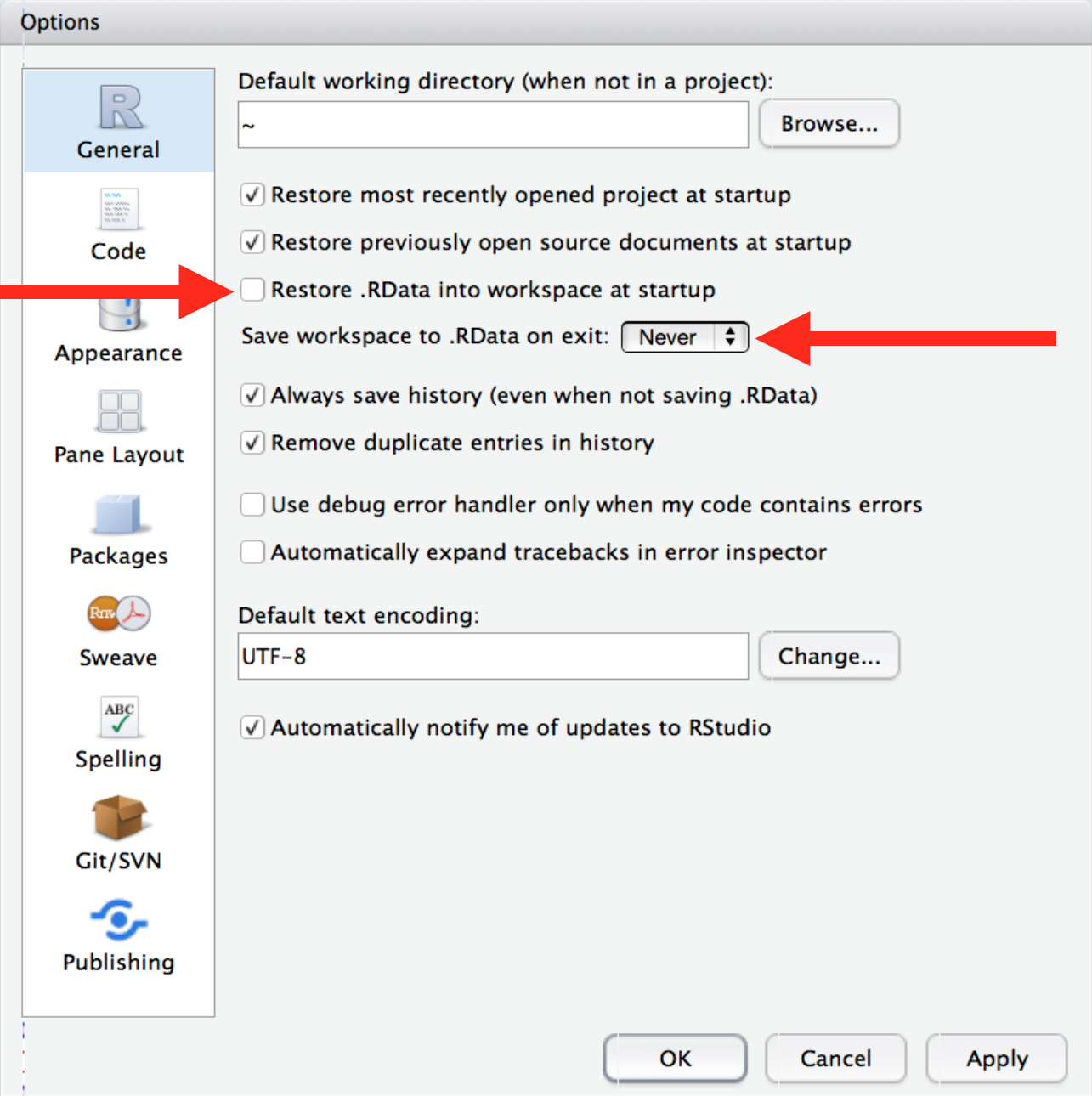
Restart R often during work
If you use RStudio, use the menu item Session > Restart R or the associated keyboard shortcut Ctrl+Shift+F10 (Windows and Linux) or Command+Shift+0 (Mac OS).
Additional keyboard shortcuts make it easy to restart development where you left off, i.e. to say “re-run all the code up to HERE”:
- In an R script, use Ctrl+Alt+B (Windows / Linux) or Command+Option+B (Mac OS)
- In R markdown, use Ctrl+Alt+P (Windows / Linux) or Command+Option+P (Mac OS)
(We’ll get back to this when talking about working with rmarkdown / quarto)
Don’t use rm(list = ls()) in your scripts
rm(list = ls()) does not remove everything in your environment (e.g. library calls, setting of working environment..)!
Caution
rm(list = ls())does not guarantee reproducibility- Instead, just make sure that you are working from a restarted R session, and don’t save your workspace
Questions for you
- How have you organized your projects in the past?
- Have you ever lost track of things? If so, how and why?
- What changes are you thinking about implementing?
File paths
Relative file paths
- Use relative paths to read in data and save stuff
Relative file paths
- Use relative paths to read in data and save stuff
Relative file paths
Tip
Use the auto-completion function in Rstudio: type ““, move the insertion point in there, and then press the tab key
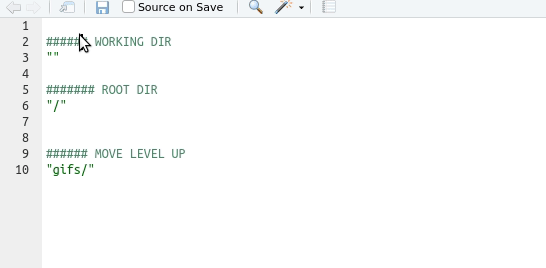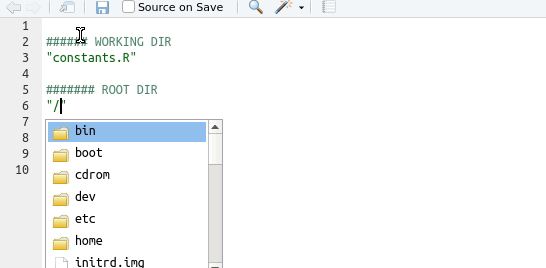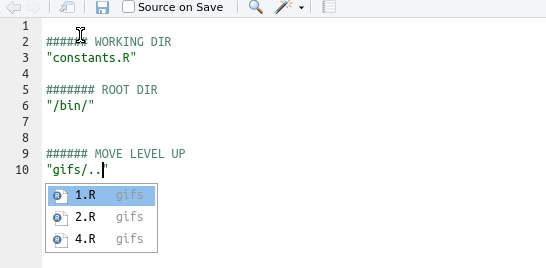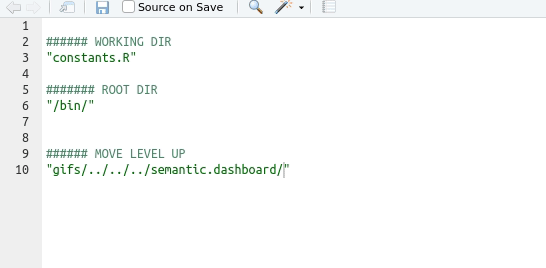
Understanding working directories
Relative file paths work because they extend on your working directory. This is one reason why you should work in an R project - it sets a working directory for you, i.e. the directory where your R project is sitting in.
The {here} package
The here() function retrieves your current working directory, at the time the package was loaded.
The {here} package helps especially when not working with R interactively (e.g. knitting an rmarkdown / quarto file)
[1] "/Users/au525642/Dropbox/postdoc/teaching & talks/23_PhD_R_course/course_materials"[1] "/Users/au525642/Dropbox/postdoc/teaching & talks/23_PhD_R_course/course_materials/data/my_data_file.csv"Compare here() with getwd()
[1] "/Users/au525642/Dropbox/postdoc/teaching & talks/23_PhD_R_course/course_materials"[1] "/Users/au525642/Dropbox/postdoc/teaching & talks/23_PhD_R_course/course_materials/Day1"The output differs because the working directory changes when the document is knitted by knitr. The here() function still produces the expected output.
Practice
- What’s your previous way to specify paths for reading and saving files?
- Try and implement the
here::here()function in an existing (or new) script- load some data using a relative path
- play around with the autocompletion